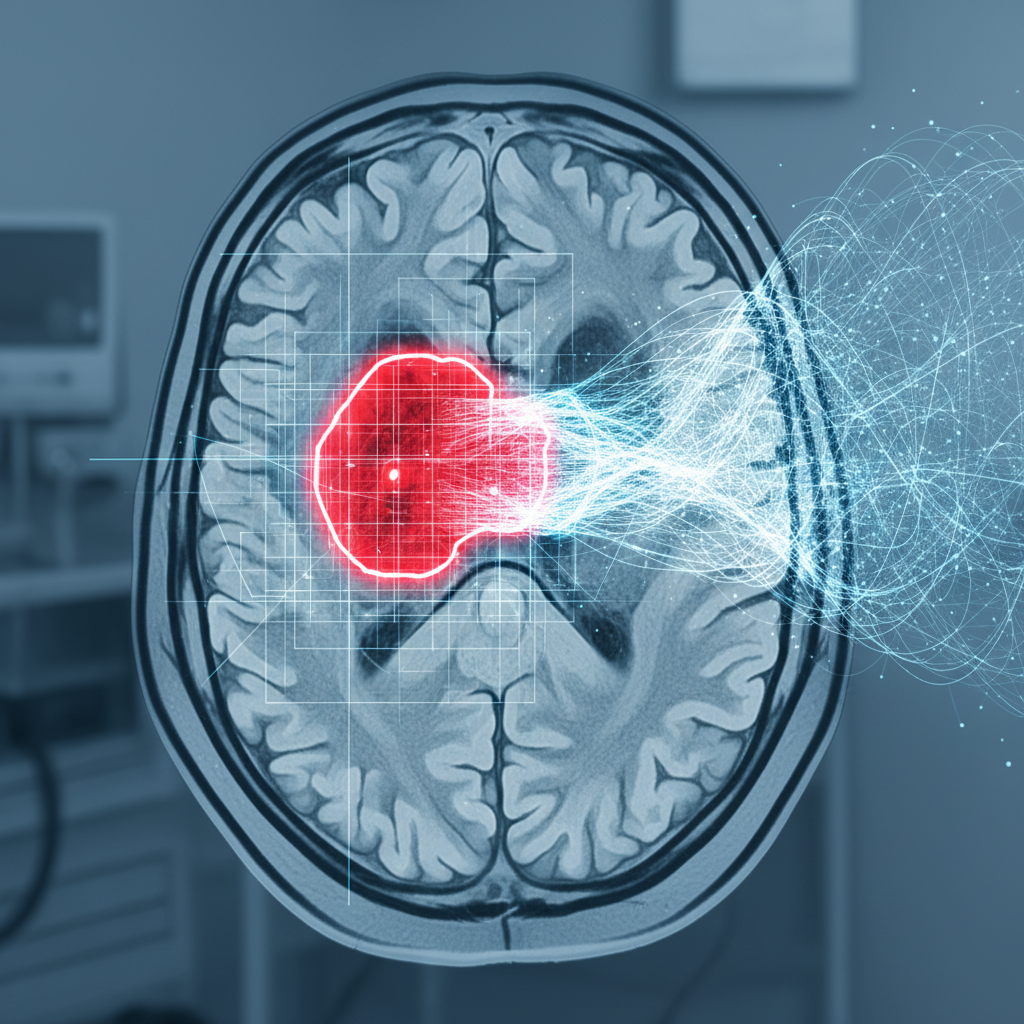
Overview
This conference poster, presented at BSEC 2011, introduces a novel computational method for predicting brain tumor growth using functional mask analysis combined with evolutionary algorithms. The research was conducted in collaboration between the University of Tennessee Machine Intelligence Lab and Oak Ridge National Laboratory’s Modeling and Simulation Group.
Key Contributions
The work presents a domain-agnostic approach to tumor growth prediction that does not rely on biological or other domain-specific knowledge about the detailed mechanisms underlying tumor growth. Instead, it leverages general systems theory principles to identify spatial-temporal patterns that have high predictive power.
Methodology
The approach consists of two main components:
-
Functional Mask Identification: Using population-based incremental learning (PBIL), an evolutionary algorithm, to efficiently search the space of possible functional masks and identify which neighboring voxels have the highest predictive power for determining future states of any given central voxel.
-
Probabilistic Parameter Estimation: Once optimal functional masks are identified, probability distributions are calculated for the next state of central voxels conditioned on the current and previous states of neighboring voxels within the mask.
Results
The experimental results demonstrated exceptional predictive accuracy:
- 98.3% accuracy over 1,300,000 predictions of next state
- 99.2% of predictions within one class of the correct class
- 95% accuracy when predicting two time steps into the future
- 96.3% of two-step predictions within one class of the correct class
Significance
This research demonstrates that general machine learning methods, when combined with systems-theoretical principles, can achieve highly accurate predictions of complex biological phenomena without requiring detailed domain knowledge. This approach has potential applications beyond brain tumor modeling, extending to any spatially-distributed dynamic system where the underlying mechanisms may be unknown or difficult to model directly.
Acknowledgements
This research was sponsored by the Laboratory Directed Research and Development Program of Oak Ridge National Laboratory, managed by UT-Battelle, LLC, for the U.S. Department of Energy. Tumor models were provided by Thomas Yankeelov and Nkiruka Atuegwu of the Vanderbilt University Institute of Imaging Science.